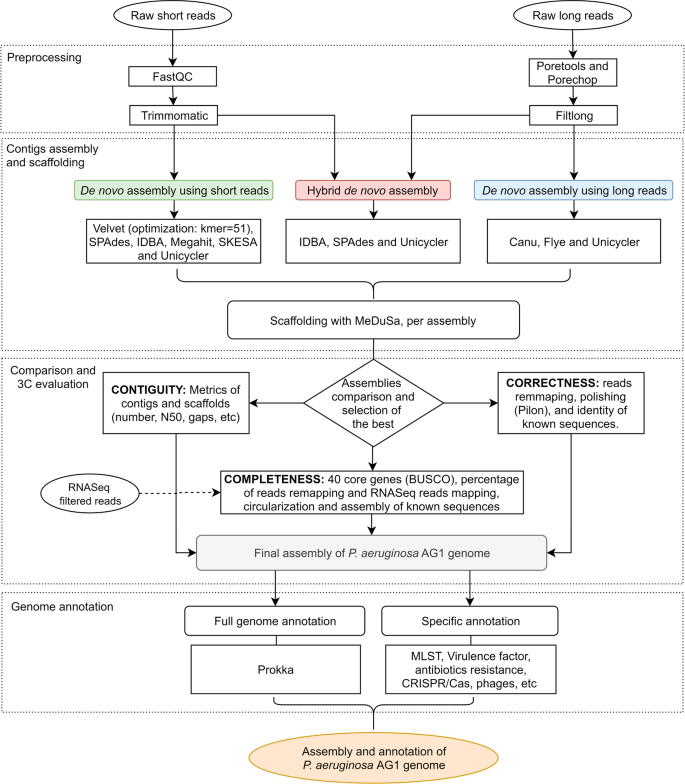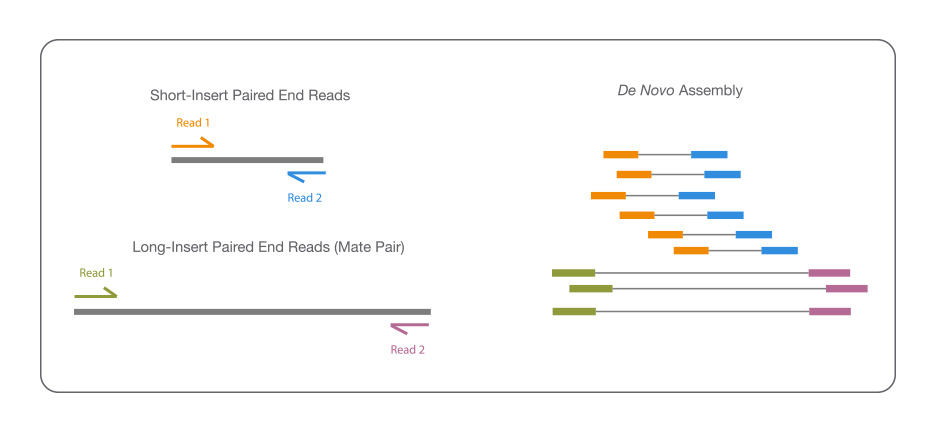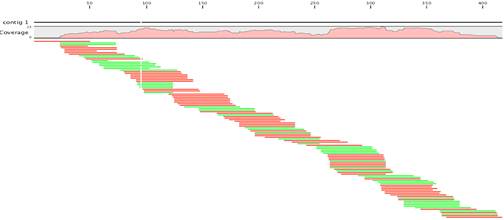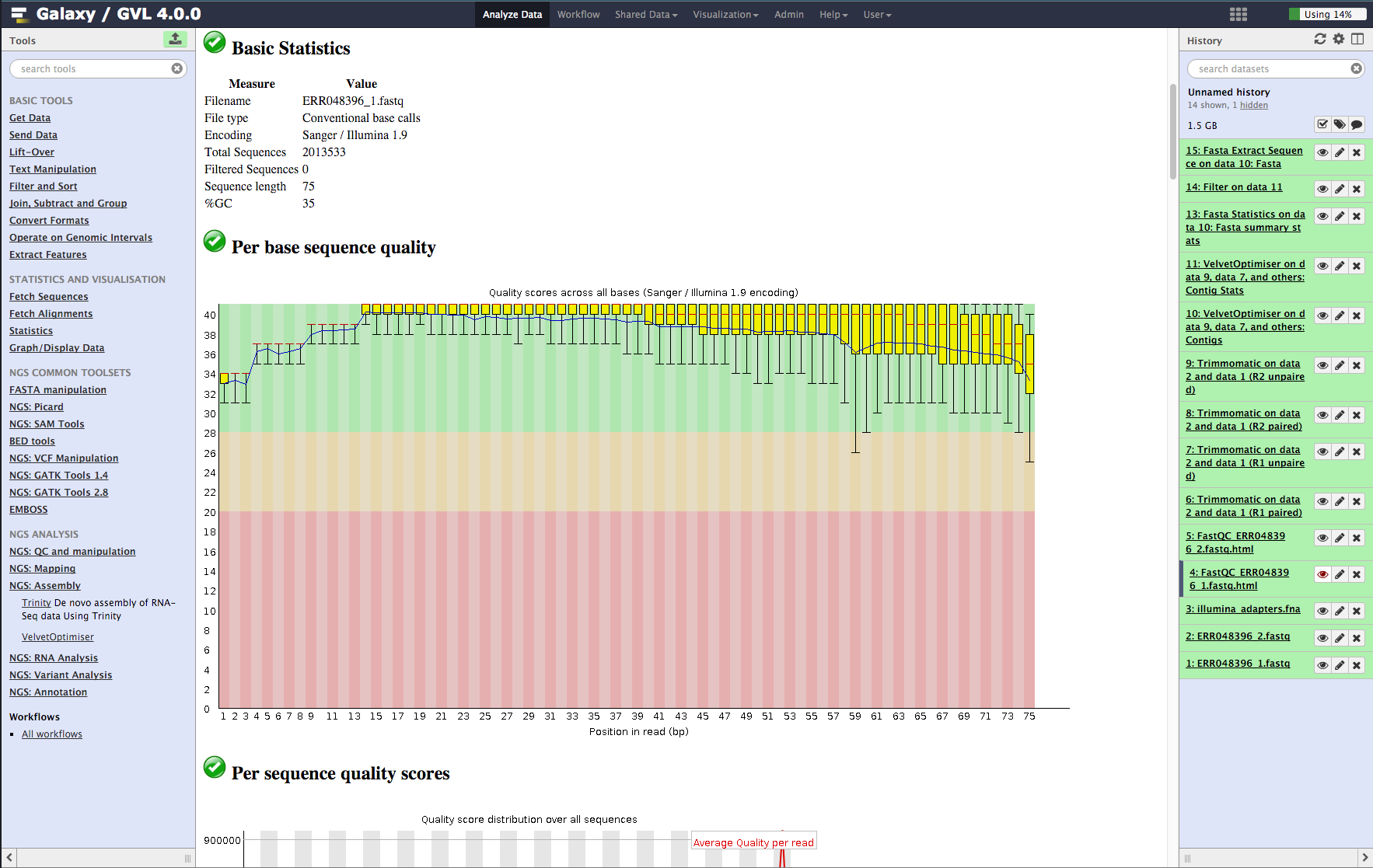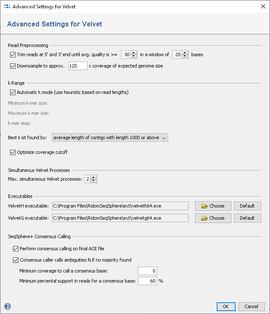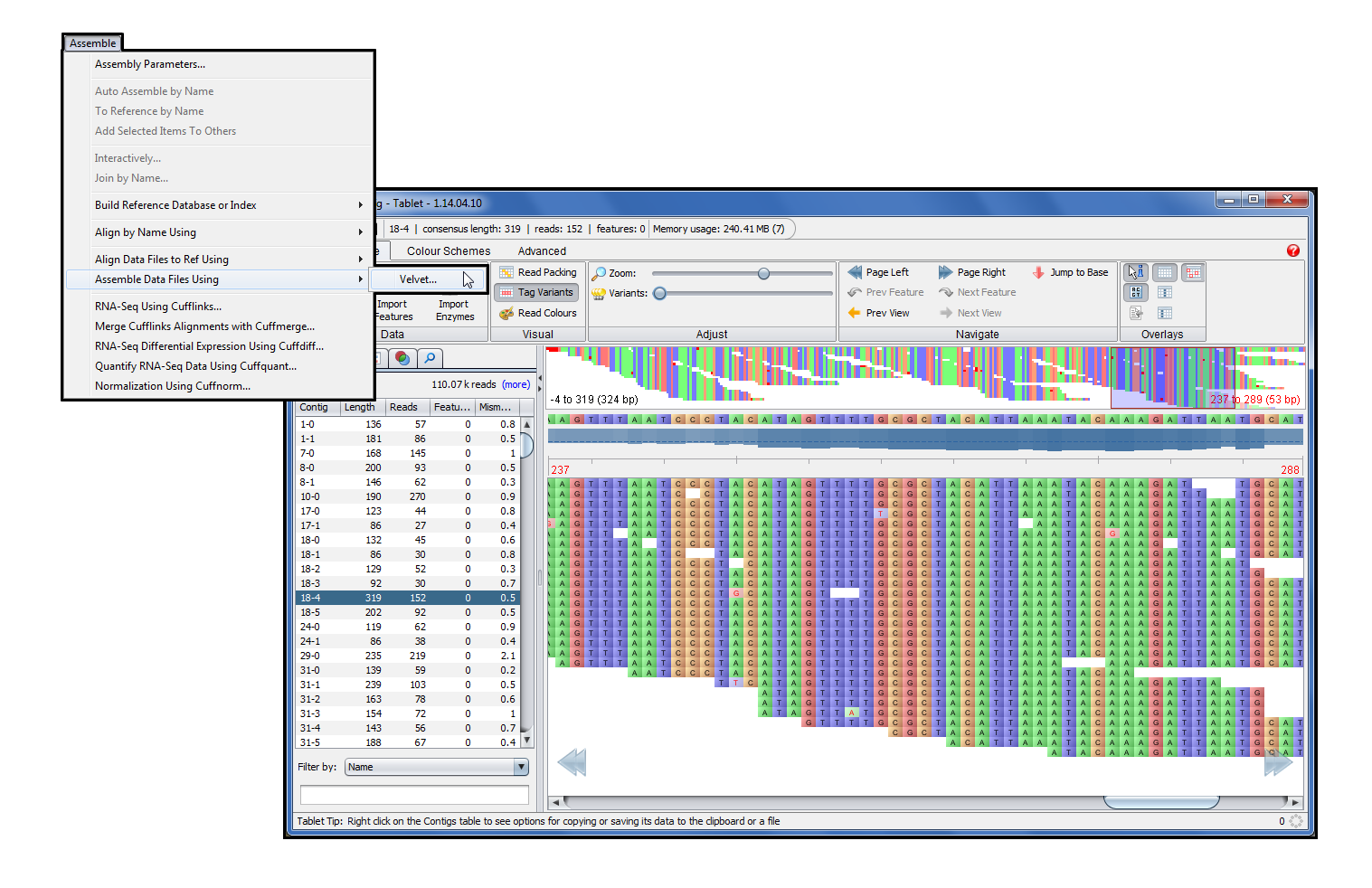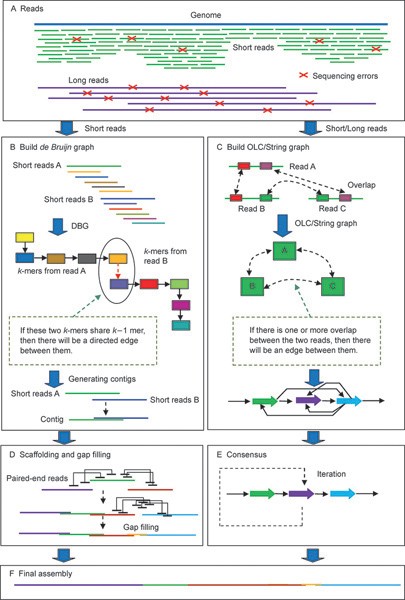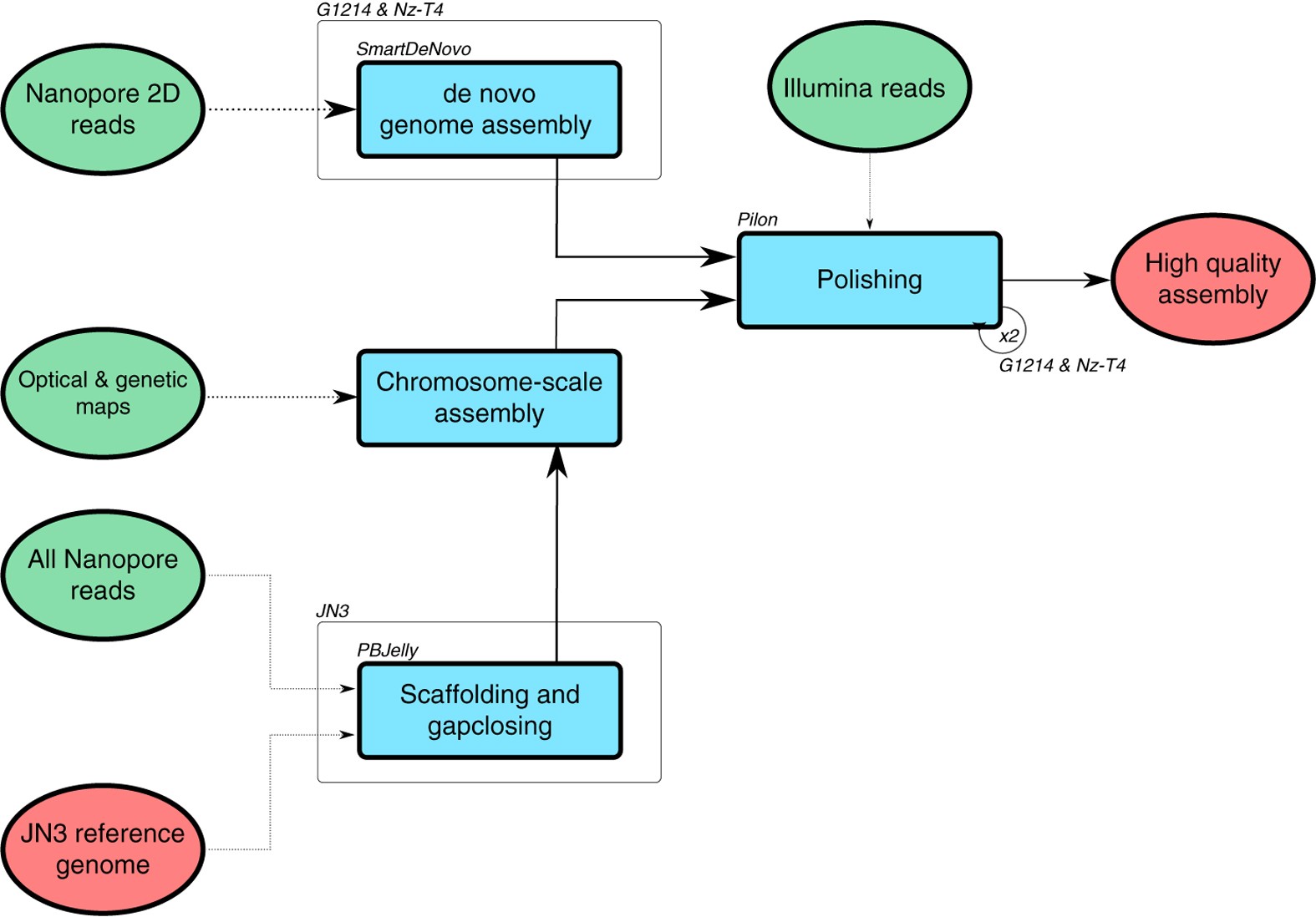
De novo assembly and annotation of three Leptosphaeria genomes using Oxford Nanopore MinION sequencing | Scientific Data
Fine De Novo Sequencing of a Fungal Genome Using only SOLiD Short Read Data: Verification on Aspergillus oryzae RIB40 | PLOS ONE

Whole Genome de novo Assembly |Supporting de novo assembly training |Course |WGS sequencing | data analysis| Workshop
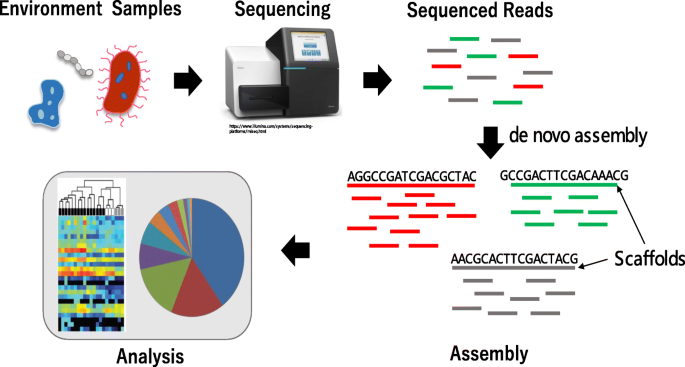
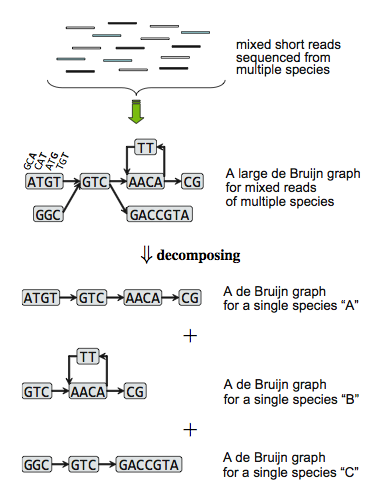
MetaVelvet-DL: a MetaVelvet deep learning extension for de novo metagenome assembly | BMC Bioinformatics | Full Text

Genome assembly with Velvet. Reads are assembled into contigs using... | Download Scientific Diagram
![PDF] MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads | Semantic Scholar PDF] MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/0de503f5ba1ca63c8b559eccb2d7dc3d8a41f862/3-Figure2-1.png)
PDF] MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads | Semantic Scholar
1 Velvet: Algorithms for De Novo Short Assembly Using De Bruijn Graphs March 12, 2008 Daniel R. Zerbino and Ewan Birney Presenter: Seunghak Lee. - ppt download
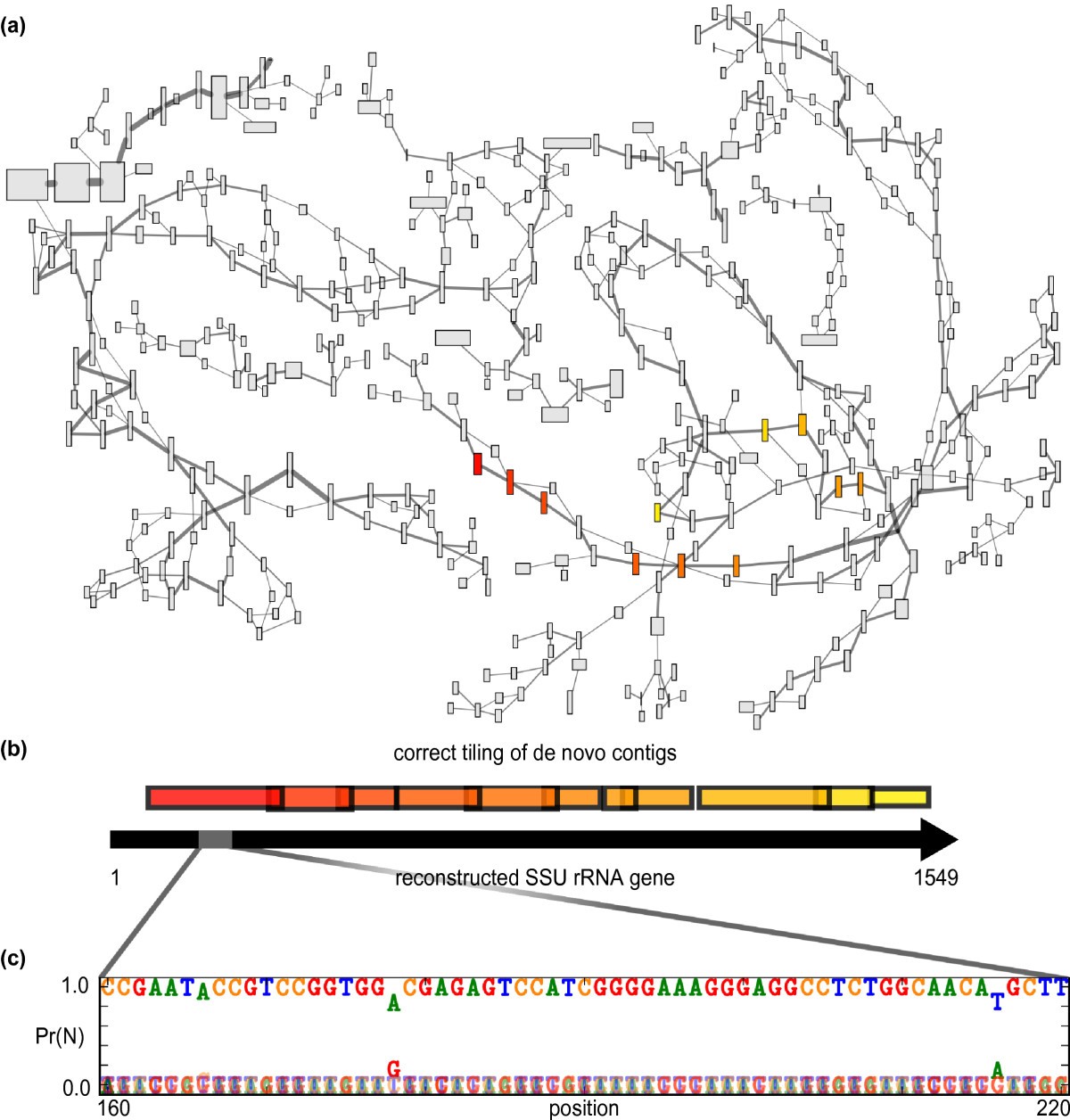
EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data | Genome Biology | Full Text

Rapid hybrid de novo assembly of a microbial genome using only short reads: Corynebacterium pseudotuberculosis I19 as a case study. | Semantic Scholar
De Novo Assembly of Bitter Gourd Transcriptomes: Gene Expression and Sequence Variations in Gynoecious and Monoecious Lines | PLOS ONE
Assembly of highly repetitive genomes using short reads: the genome of discrete typing unit III Trypanosoma cruzi strain 231

Summary of iterative de novo assembly using Velvet. Boxes with borders... | Download Scientific Diagram